After obtaining a Bachelor's degree in Human Biology, I pursued a Master's degree in Bioinformatics. I am passionate about software engineering, web development, and data analysis applied to biology. I am eager to apply the skills I acquired during my studies and professional experiences in a position in bioinformatics.
This website is one of my personal projects. It showcases my programming skills, particularly in HTML and CSS, and is available in French. I have included links to the various training programs I participated in, and you can contact me through my LinkedIn profile or by email.

Python, Java, C, C++, Bash
HTML, CSS, JavaScript, TypeScript, Angular
PostgreSQL
Linux, MacOS, Windows
Design Patterns, XML, JSON, UML
BLAST, Galaxy, Bowtie2, ClustalW
Data Mining, R Programming Language
ImageJ
Visualization of structural data in 2D and 3D (SVG and WebGL)
Human Biology, Pathophysiology, Biochemistry
The European Molecular Biology Laboratory (EMBL) includes the European Bioinformatics Institute (EBI) among its establishments. The aim of EBI is to generate and centralise molecular data to make it accessible to the public. I had the opportunity to work within the team that deals with molecular networks. During my internship at the organisation, I had the chance to develop the Molecular Complex Navigator, a web tool for visualising and comparing protein complexes, available by clicking on this link. This web tool, developed in TypeScript with the Angular framework, is both performant and interactive. The integration of data such as orthology allows for the comparison of complexes across species. This required the addition of new references to the Panther database, integrated into our own database using code in Java utilising the Spring framework. This project allowed me to develop my skills in web development and communication, particularly through the various presentations I delivered (Scientific Advisory Board, EMBL-EBI Protein and Complexes Day, HUPO Conference 2024).
Following my internship at EMBL-EBI, I had the opportunity to remain in the molecular networks team to update and enhance the XML Maker. This Java application, using the Spring framework, is an essential tool for scientific curators, allowing them to generate XML files from data provided by laboratories. The first stage of this project was the implementation of a formatter that enables curators to convert gene and protein names into UniProtKB accession numbers. This represents a considerable time-saving for curators who previously extracted this data manually. The second stage of this project, which is still ongoing, is the redesign of the XML file generator to implement the standard PSI-MI formats using an appropriate library.
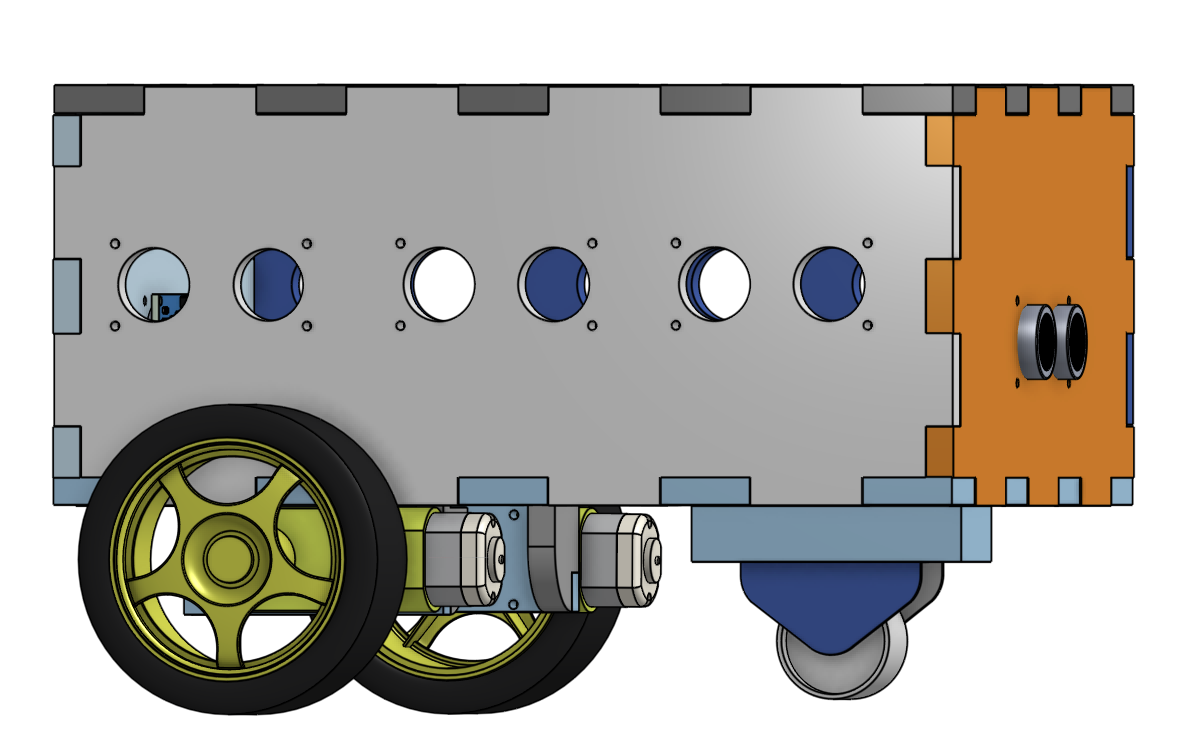
The objective of this project was to construct a fleet of robots detected by ArUco markers via an external camera. As part of this group project, we modelled the robot using OnShape software, and then we assembled the robots. These robots are equipped with an ESP32 microcontroller and an L298N driver that allows control of two motors.
My contribution to the project was the implementation of code on the microcontroller via the Arduino IDE (Integrated Development Environment). This IDE uses the C++ programming language, which is geared towards hardware programming. I also developed the website to control the robots, using HTML.